10. Spatial Relation Of Genomic (SROG) intervals
10.1. Description
Match up two sets of genomic intervals, and report the code of Spatial Relation Of Genomic (SROG).
SROG codes include disjoint, touch, equal, overlap, contain, within.
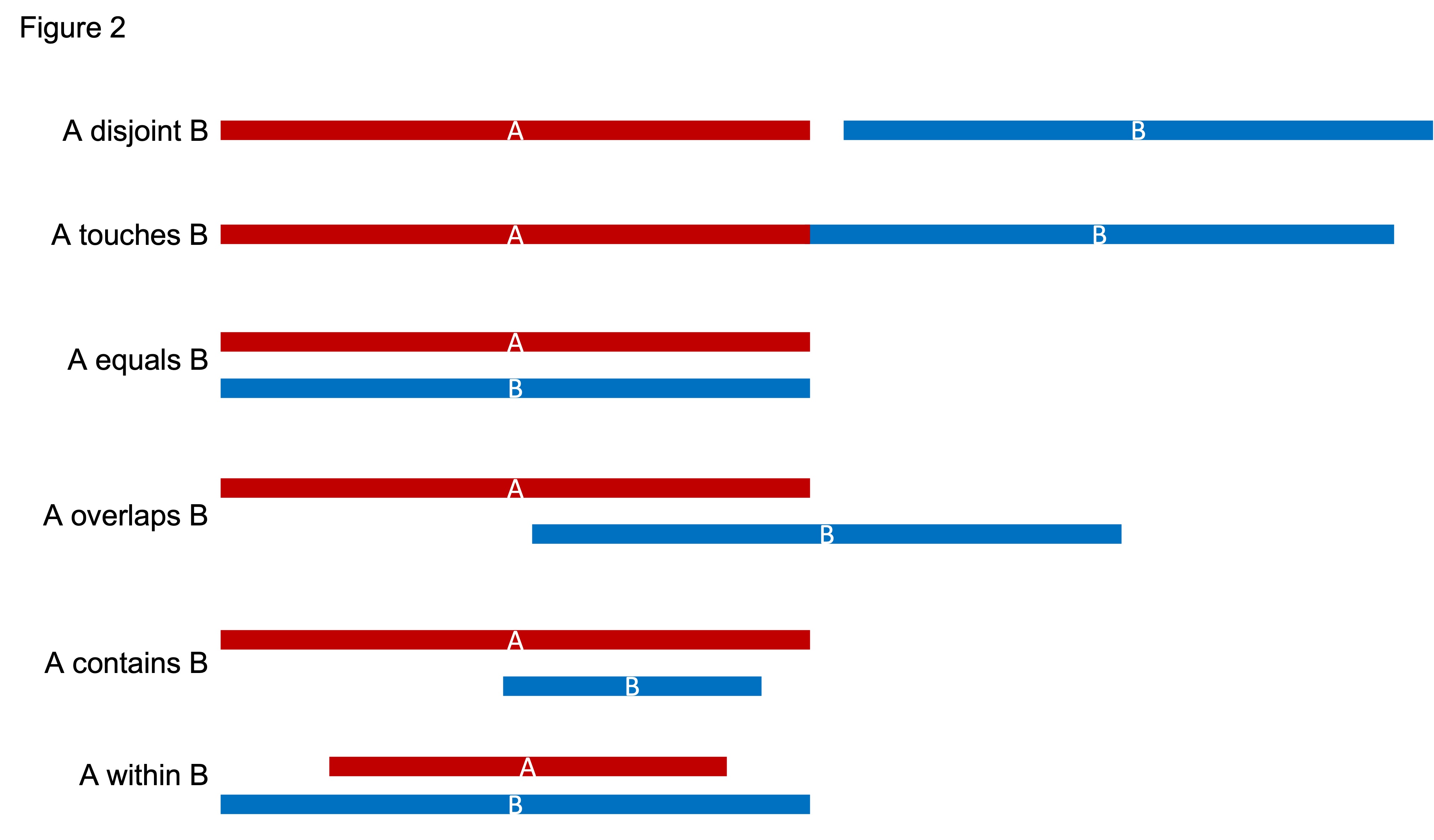
10.2. Usage
cobind.py srog -h
usage: cobind.py srog [-h] [--dist MAX_DIST] [-l log_file] [-d]
input_A.bed input_B.bed output.tsv
positional arguments:
input_A.bed Genomic regions in BED, BED-like or bigBed format. If
'name' (the 4th column) is not provided, the default
name is "chrom:start-end". If strand (the 6th column)
is not provided, the default strand is "+".
input_B.bed Genomic regions in BED, BED-like or bigBed format. If
'name' (the 4th column) is not provided, the default
name is "chrom:start-end". If strand (the 6th column)
is not provided, the default strand is "+".
output.tsv Generate spatial relation code (disjoint, touch,
equal, overlap, contain, within) for each genomic
interval in "input_A.bed".
options:
-h, --help show this help message and exit
--dist MAX_DIST When intervals are disjoint, find the closest up- and
down-stream intervals that are no further than
`max_dist` away. default: 250000000)
-l log_file, --log log_file
This file is used to save the log information. By
default, if no file is specified (None), the log
information will be printed to the screen.
-d, --debug Print detailed information for debugging.
10.3. Example
cobind.py srog CTCF_ENCFF660GHM.bed3 RAD21_ENCFF057JFH.bed3 output.tsv
2022-01-20 09:01:17 [INFO] Determine the spacial realtions of genomic (SROG) intervals ...
2022-01-20 09:01:17 [INFO] Build interval tree from file: "RAD21_ENCFF057JFH.bed3"
2022-01-20 09:01:17 [INFO] Reading BED file: "CTCF_ENCFF660GHM.bed3"
disjoint 30419
overlap 4341
contain 1695
within 23214
touch 0
equal 1
other 0
dtype: int64
Match up results were saved to output.tsv
$head -10 output.tsv
chr12 53676079 53676369 within chr12:53676060-53676382
chr12 57905364 57905661 within chr12:57905272-57905699
chr22 20564334 20564661 contain chr22:20564370-20564581
chr16 57649065 57649362 within chr16:57649007-57649370
chr17 45135294 45135610 overlap chr17:45135296-45135642
chr15 40274737 40275016 within chr15:40274714-40275018
chr1 114346538 114346847 within chr1:114346526-114346903
chr7 151172578 151172888 overlap chr7:151172565-151172865
chr1 225474965 225475268 within chr1:225474919-225475330
chr5 179668464 179668730 contain chr5:179668495-179668674
...
chr22 23128466 23128723 disjoint UpInterval=chr22:22651059-22651463,
DownInterval=chr22:23385972-23386169
...
- Column 1-3
Genome intervals from “CTCF_ENCFF660GHM.bed3”.
- Column 4
SROG code. When SORG =
disjoint, two closest intervals (up- and down-stream) fromRAD21_ENCFF057JFH.bed3were reported.- column 5
Genomic intervals from
RAD21_ENCFF057JFH.bed3.